Methods in population genomics
Ongoing project
There are many methods in population genomics. To help newcomers, we published a review on the main methods in the field, available here
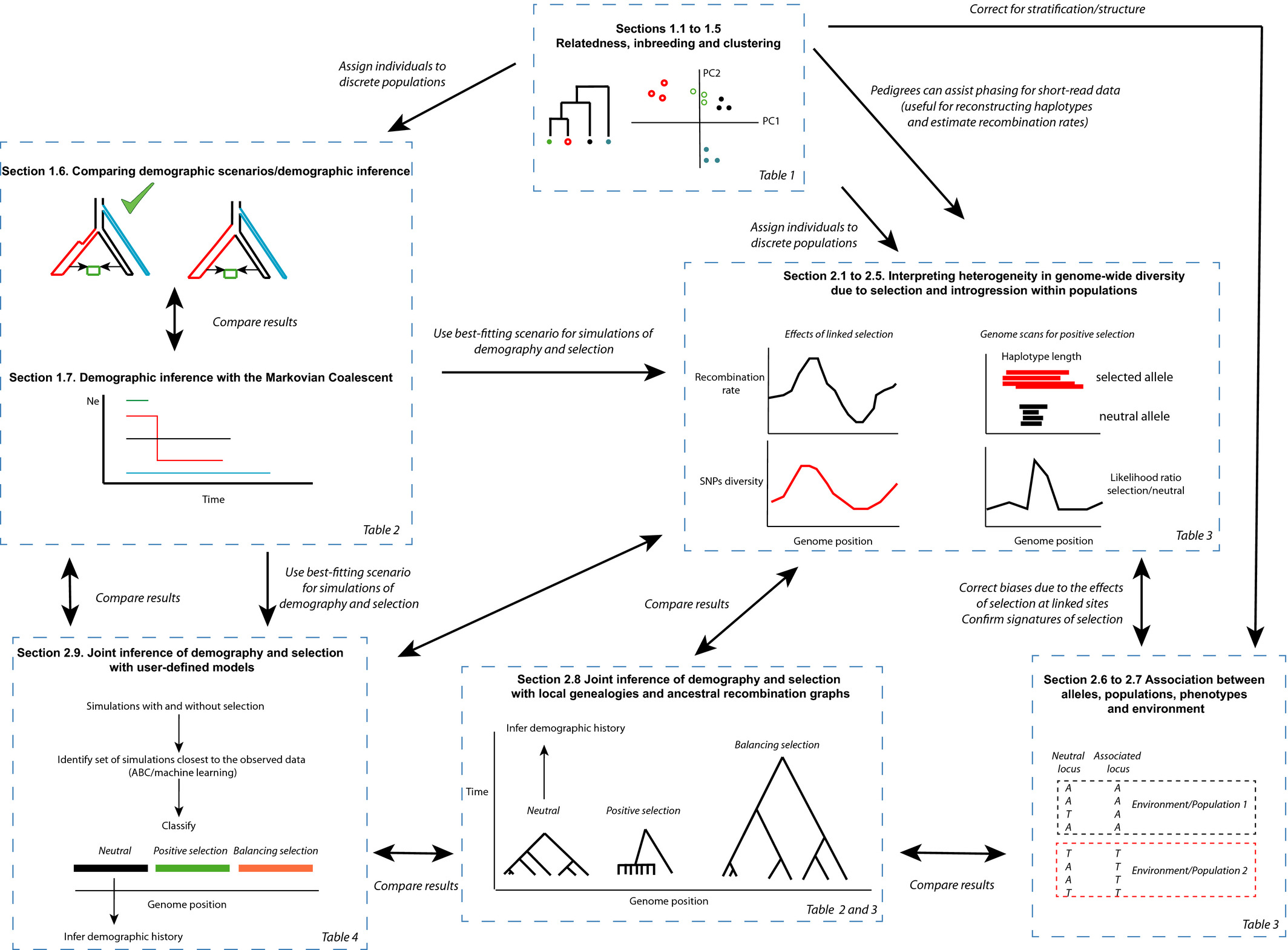
This simple graph should explain it all…
A few more resources.
I try to regularly update the tables so they reflect the most recent methods. You can also check the blog posts, where I pin reviews or articles that summarize recent methods.
I also design lectures and workshops in population genomics. I recently contributed with Thibault Leroy to an online Physalia course.
In collaboration with Julie Orjuela at IRD, we designed a three-days workshop on population genomics. It is inspired by the one designed for Physalia, but also includes new material, particularly for demographic inference. Data for workshops #1 and #2 can be found at this address
Workshop #1: Population structure
Workshop #2: Demographic inference, introduction to ABC and likelihood approaches
Workshop #3: Selection and association scans
I am also planning another set of workshops on spatial genomics. I recently started developing pipelines using slendr, which is a R package interacting with tskit and SLiM, and can be very useful if your research has a spatial component and you want to test the robustness of your analyses. I will post it here once done.
Table 1: Summary of methods dedicated to infer population structure.
| Software | Class of method | Purpose | Class of method | Issues/Warnings | Reference | Link |
|---|
Table 2: Summary of methods dedicated to detect selection
| Software | Class of method | Purpose | Class of method | Issues/Warnings | Reference | Link |
|---|
Table 3: Summary of methods dedicated to reconstruct demographic history
| Software | Class of method | Purpose | Class of method | Issues/Warnings | Reference | Link |
|---|
Table 4: Summary of simulators and simulation-based methods for selection and demographic inference.
| Software | Class of method | Purpose | Class of method | Issues/Warnings | Reference | Link |
|---|